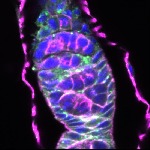
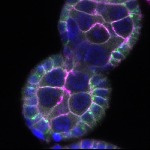
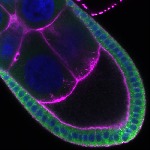
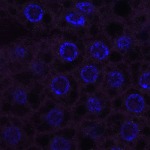
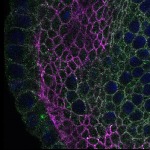
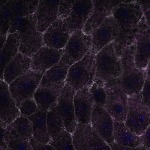
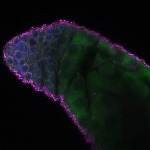
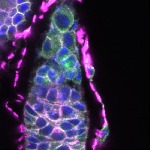
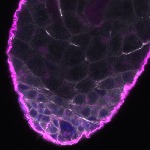
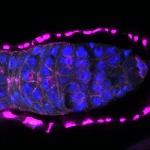
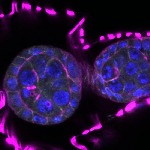
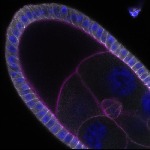
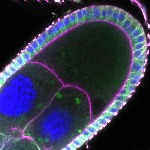
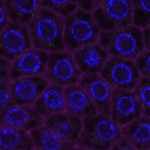
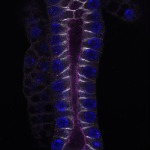
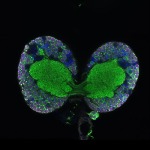
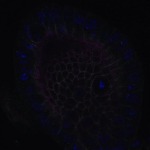
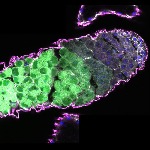
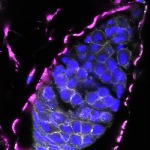
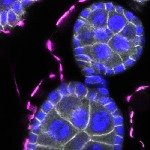
Welcome to FLYtRAB
The database to explore Drosophila Rab protein cell type expression and subcellular localization across diverse tissues.
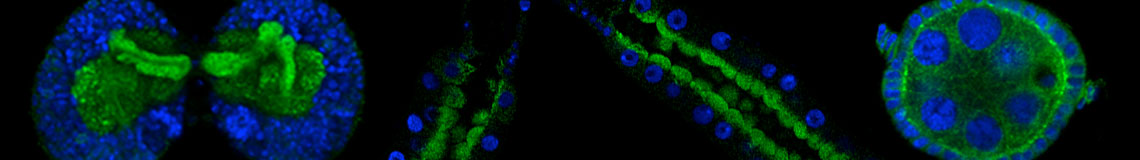
Rab proteins in Fruit flies
DNA sequence analysis identified 33 genes [1, 2, 3] encoding Rab proteins in Drosophila melanogaster. 23 of these are orthologues of vertebrate Rabs [1, 3, 4] and almost all give rise to only one protein isoform [23] Rabs control specific steps in intracellular lipid and protein traffic [5, 6, 7]. Hence, Rabs represent not only valuable identification marks for organelles [8, 9, 10, 11, 12] and membrane compartments [13, 14, 15, 16, 17], but serve as logical targets for manipulation of specific intracellular transport routes [18, 19, 20, 21].
The endogenous YFP-Rab-Fly stock collection
We have established a genetic resource that allows systematic, direct and controlled access to the Rab machinery in Drosophila melanogaster. Using homologous recombination, we targeted all functional genomic rab loci, fusing an YFP4xMyc tag to the N-terminal end of each Rab protein. The resultant rab alleles are viable and do not show obvious phenotypes. The common YFP-tag enables systematic and quantitative Rab mRNA and protein detection in vivo, and available genetic knock-down tools directed against GFP efficiently reduce YFP-Rab protein levels.
Available open source data
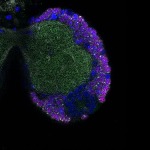
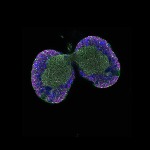
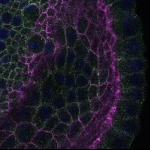
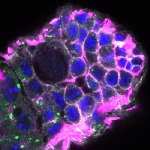
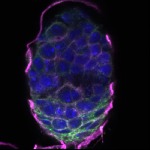
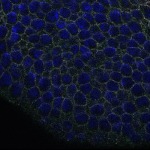
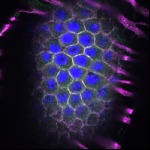
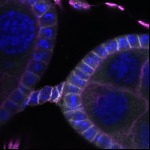
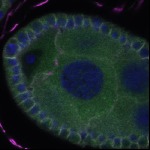
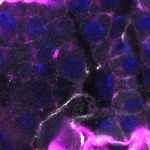
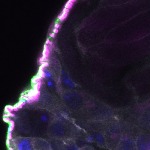
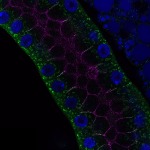
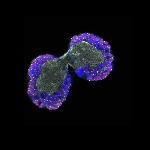
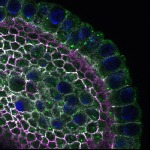
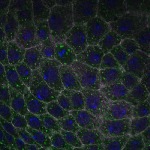
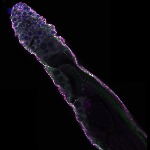
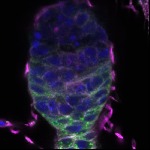
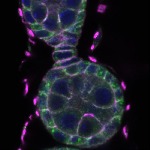
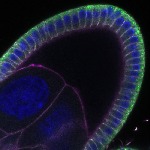
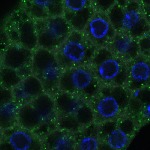
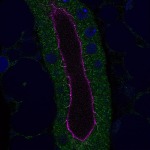
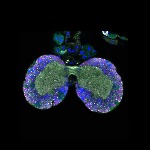
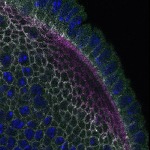
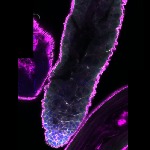
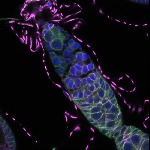
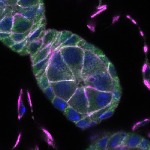
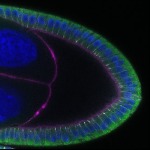
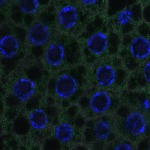
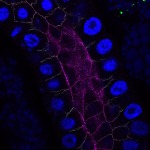
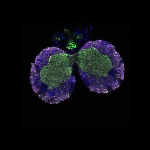
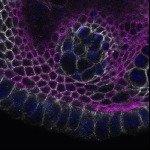
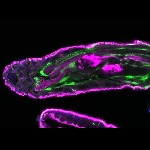
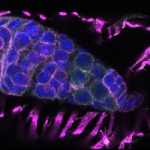
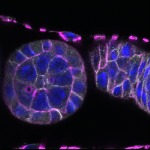
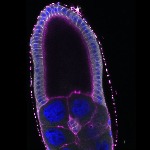
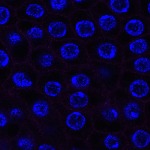
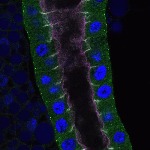
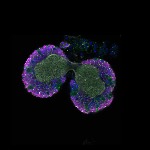
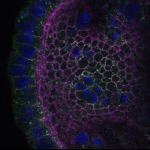
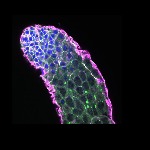
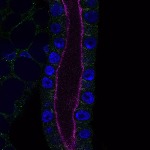
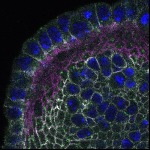
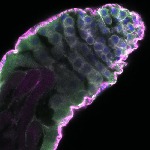
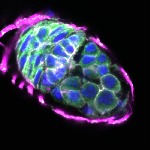
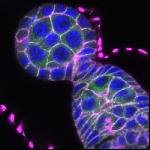
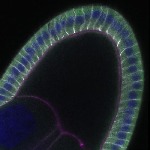
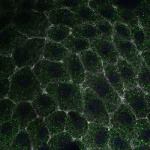
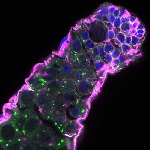
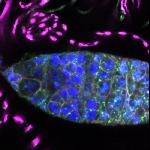
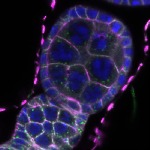
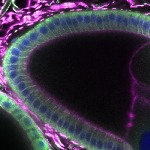
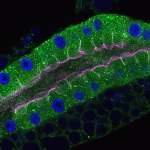
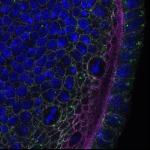
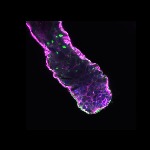
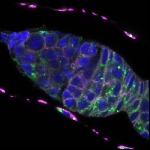
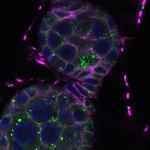
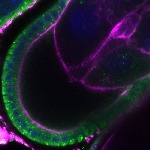
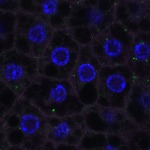
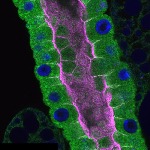
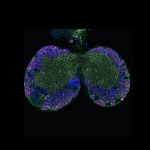
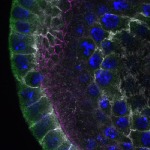
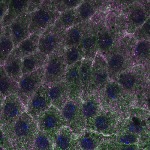
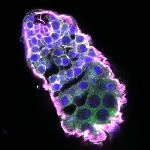
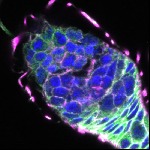
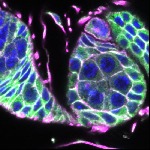
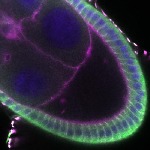
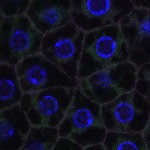
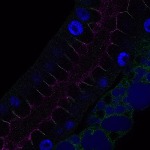
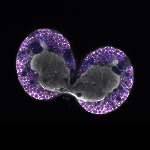
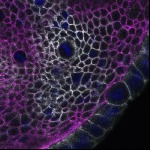
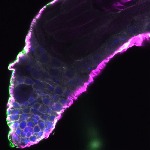
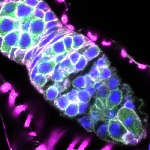
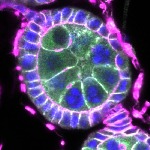
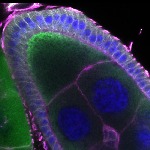
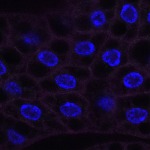
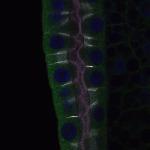
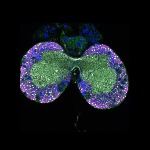
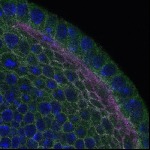
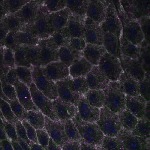
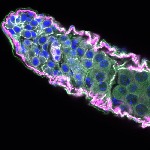
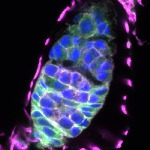
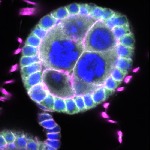
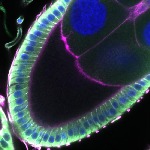
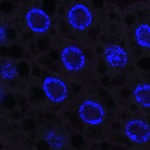
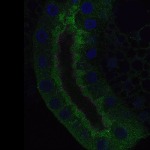
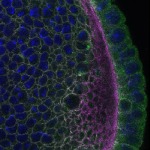
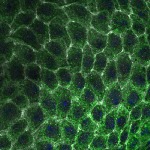
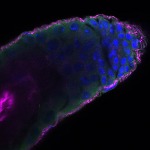
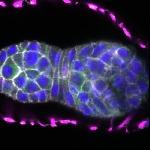
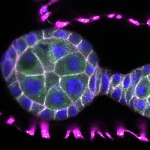
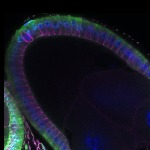
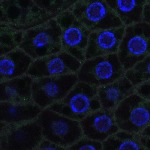
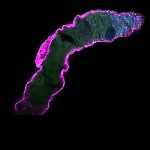
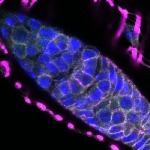
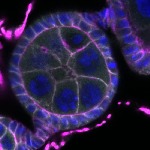
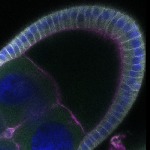
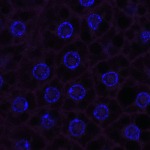
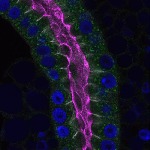
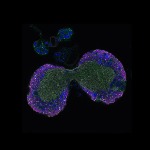
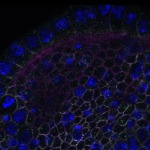
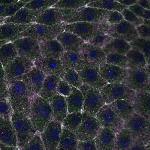
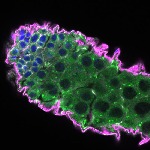
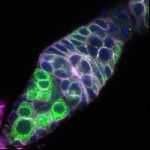
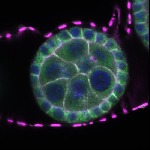
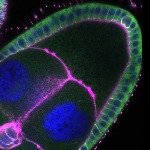
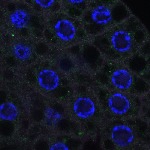
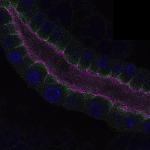
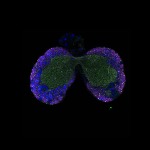
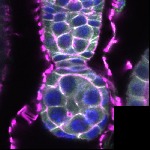
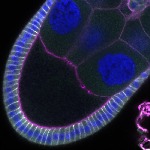
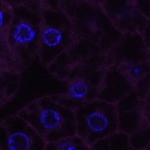
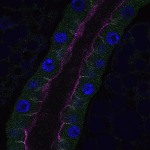
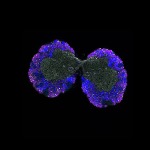
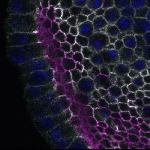
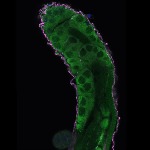
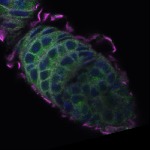
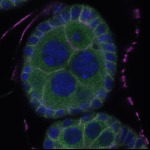
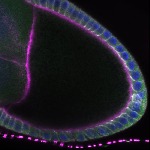
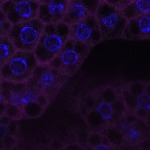
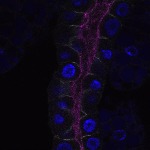
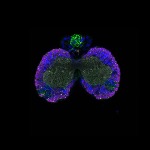
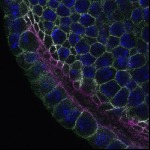
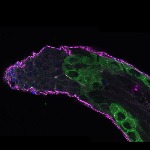
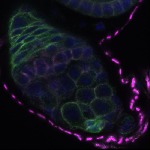
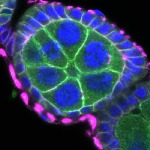
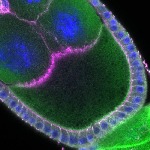
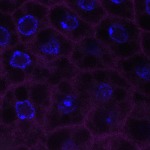
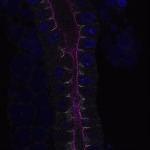
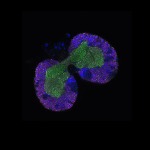
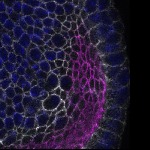
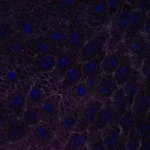
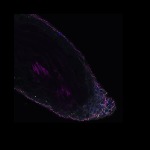
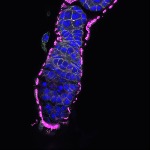
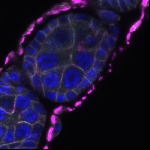
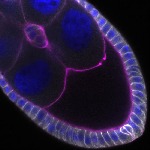
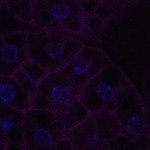
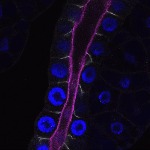
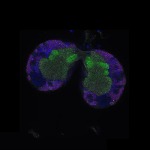
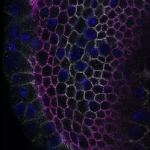
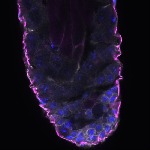
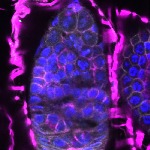
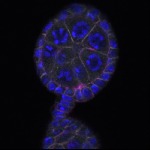
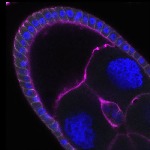
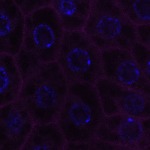
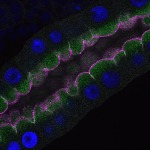
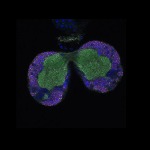
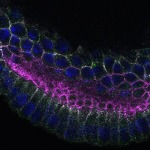
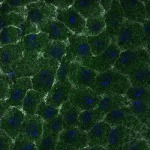
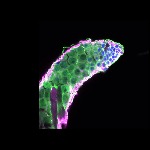
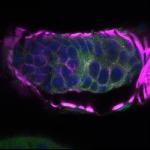
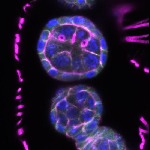
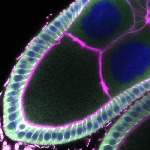
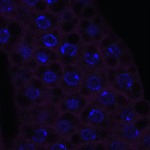
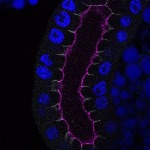
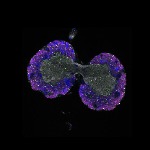
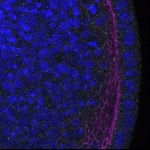
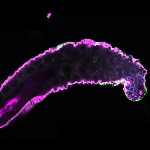
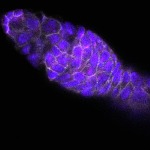
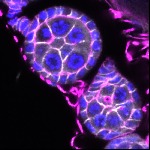
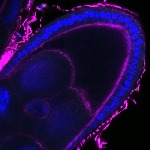
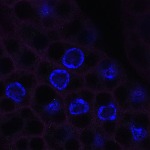
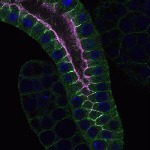
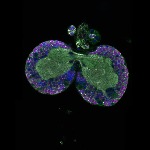
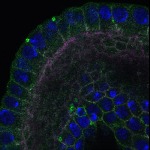
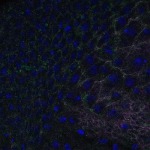
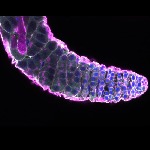
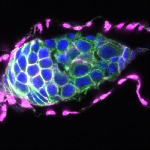
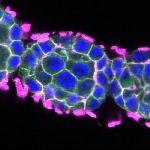
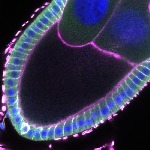
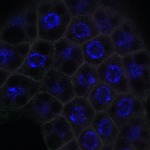
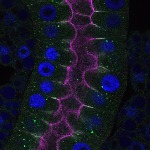
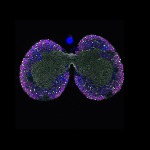
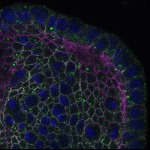
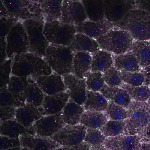
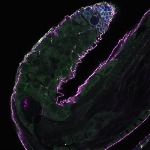
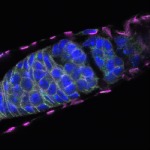
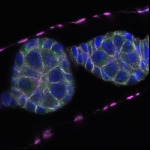
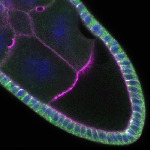
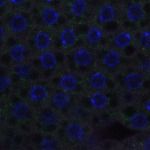
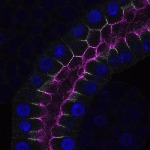
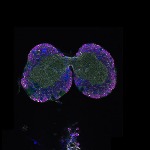
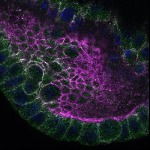
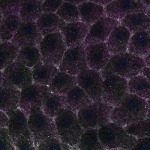
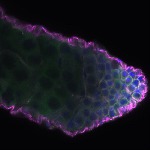
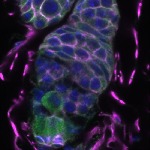
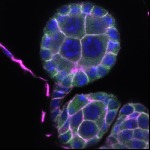
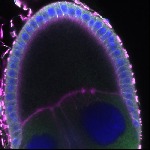
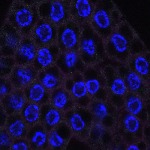
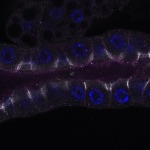
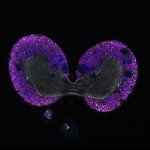
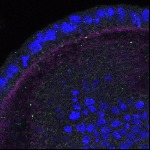
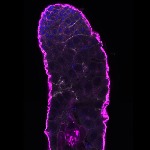
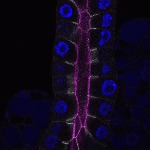
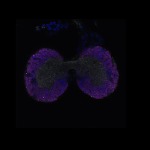
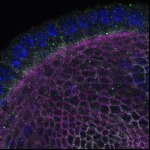
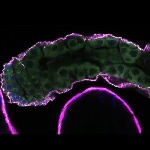
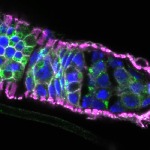
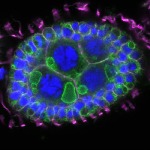
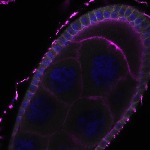
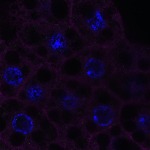
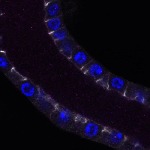
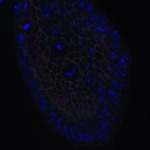
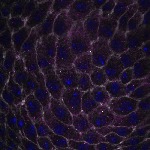
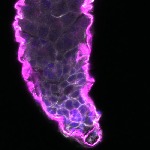
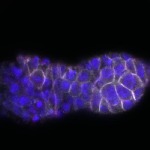
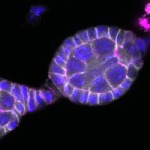
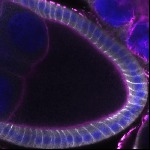
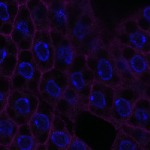
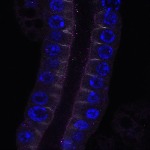
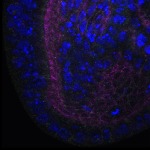
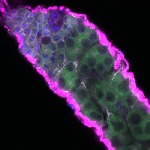
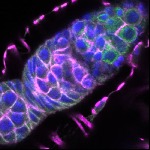
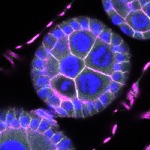
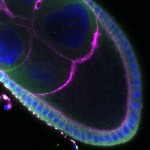
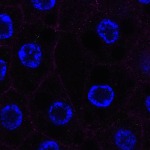
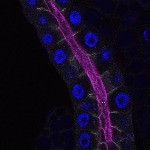
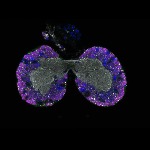
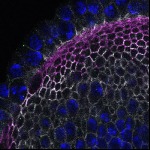
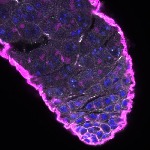
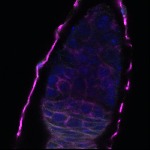
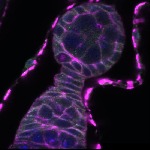
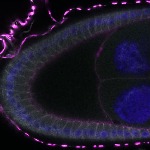
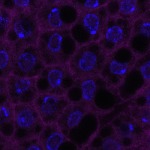
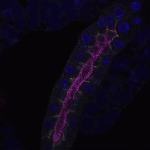
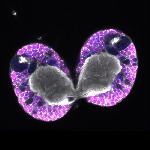
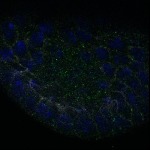
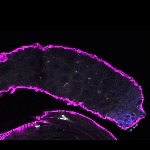
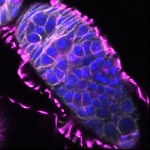
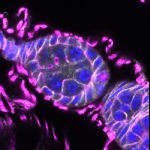
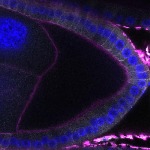
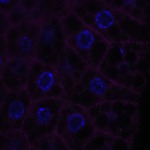
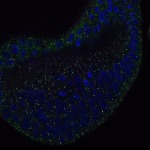
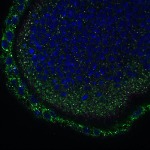
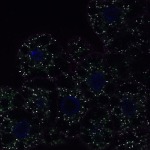
We imaged all YFP-Rab proteins with subcellular resolution in larval Fat bodies, Salivary glands, Wing discs, CNS and adult Ovaries and Testes [24, 25]. These organs compromise over 20 different cell types and we used controlled vocabulary to annotate the acquired images, describing features such as tissue and cell type specific expression, morphology and subcellular localisation.
The open access FLYtRAB database provides direct access to CATMAID [22], a free online platform that enables the user to view and download all 3D image data from the FLYtRAB library. In addition, FLYtRAB enables users to search for annotation terms linked to stored image data.
References
- [1]
- J. Zhang et al., Thirty-one flavors of Drosophila rab proteins. Genetics 176, 1307 (Jun, 2007).
- [2]
- C. C. Chan et al., Systematic discovery of Rab GTPases with synaptic functions in Drosophila. Curr Biol 21, 1704 (Oct 25, 2011).
- [3]
- Y. Diekmann et al., Thousands of rab GTPases for the cell biologist. PLoS Comput Biol 7, e1002217 (Oct, 2011).
- [4]
- J. B. Pereira-Leal, M. C. Seabra, Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313, 889 (Nov 2, 2001).
- [5]
- H. Stenmark, Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10, 513 (Aug, 2009).
- [6]
- S. R. Pfeffer, Rab GTPase regulation of membrane identity. Curr Opin Cell Biol 25, 414 (Aug, 2013).
- [7]
- D. F. Markgraf, K. Peplowska, C. Ungermann, Rab cascades and tethering factors in the endomembrane system. FEBS Lett 581, 2125 (May 22, 2007).
- [8]
- S. R. Pfeffer, Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol 11, 487 (Dec, 2001).
- [9]
- R. Behnia, S. Munro, Organelle identity and the signposts for membrane traffic. Nature 438, 597 (Dec 1, 2005).
- [10]
- T. Gronemeyer et al., Localization of Rab proteins to peroxisomes: a proteomics and immunofluorescence study. FEBS Lett 587, 328 (Feb 14, 2013).
- [11]
- C. O. Sandoval, T. Simmen, Rab proteins of the endoplasmic reticulum: functions and interactors. Biochem Soc Trans 40, 1426 (Dec 1, 2012).
- [12]
- R. Sinka, A. K. Gillingham, V. Kondylis, S. Munro, Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol 183, 607 (Nov 17, 2008).
- [13]
- M. Zerial, H. McBride, Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2, 107 (Feb, 2001).
- [14]
- B. L. Grosshans, D. Ortiz, P. Novick, Rabs and their effectors: achieving specificity in membrane traffic. Proceedings of the National Academy of Sciences of the United States of America 103, 11821 (Aug 8, 2006).
- [15]
- M. C. Seabra, C. Wasmeier, Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol 16, 451 (Aug, 2004).
- [16]
- M. Cabrera, C. Ungermann, Guanine nucleotide exchange factors (GEFs) have a critical but not exclusive role in organelle localization of Rab GTPases. J Biol Chem 288, 28704 (Oct 4, 2013).
- [17]
- F. A. Barr, Review series: Rab GTPases and membrane identity: causal or inconsequential? J Cell Biol 202, 191 (Jul 22, 2013).
- [18]
- F. Schimmoller, I. Simon, S. R. Pfeffer, Rab GTPases, directors of vesicle docking. J Biol Chem 273, 22161 (Aug 28, 1998).
- [19]
- M. C. Seabra, E. Coudrier, Rab GTPases and myosin motors in organelle motility. Traffic 5, 393 (Jun, 2004).
- [20]
- Y. Jin et al., Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev Cell 21, 1156 (Dec 13, 2011).
- [21]
- T. Bhuin, J. K. Roy, Rab proteins: The key regulators of intracellular vesicle transport. Exp Cell Res, (Aug 1, 2014).
- [22]
- S. Saalfeld, A. Cardona, V. Hartenstein, P. Tomancak, CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25, 1984 (Aug 1, 2009).
- [23]
- Flybase (http://flybase.org)
- [24]
- Flybase Atlas (http://flybase.org/static_pages/imagebrowser/imagebrowser10.html)
- [25]
- Interactive Fly (http://www.sdbonline.org/sites/fly/aimain/1aahome.htm)